When working with NEBNext Multiplex Oligos, please confirm the orientation of your indices when setting up the sample sheet for G4 sequencing.
INTRODUCTION
NEBNext Multiplex Oligos are compatible with the G4 Sequencing Platform adaptors to enable high-yield multiplex libraries. Due to the G4 index sequencing directionality and index length requirements, we recommend a few modifications to how the NEBNext Index sequences are entered in a Singular Genomics (SG) Compatible Sample Sheet relative to sample sheets for other Third-party sequencing platforms.
REVERSE COMPLEMENTING THE INDEX SEQUENCES
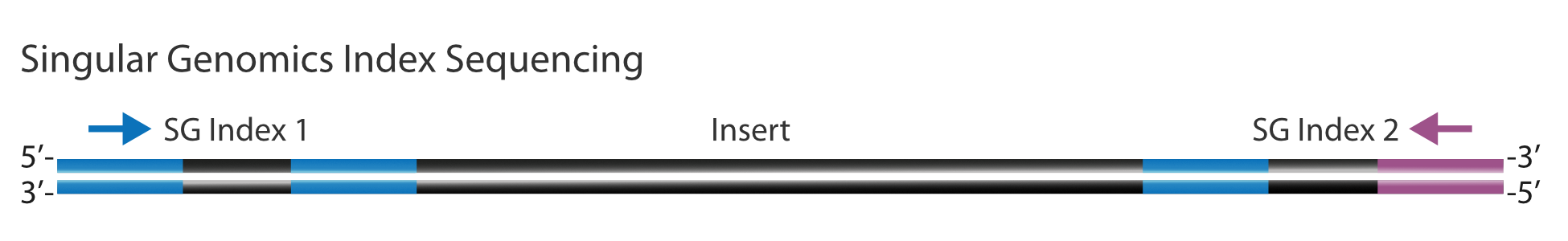
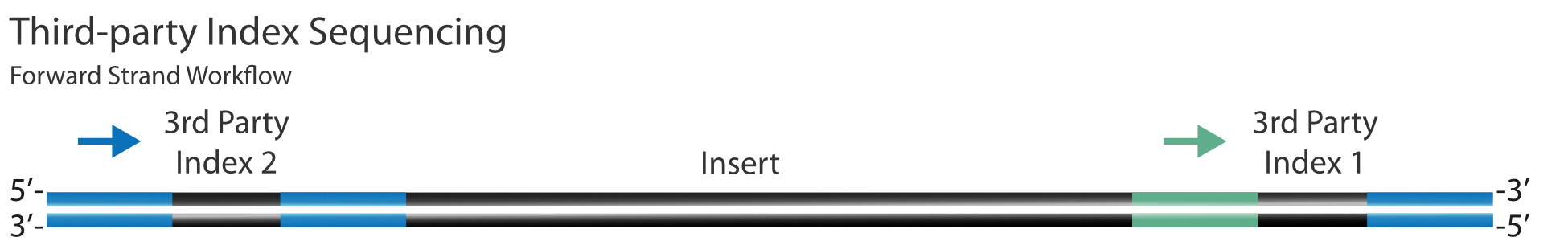
The G4 Sequencing Platform sequences the index reads from the flow cell binding sequences, S1 and S2 (see diagram below). The SG Index 2 is sequenced in the orientation shown by the purple arrow below; however, on other Third-party sequencing platforms, it will typically be sequenced in the opposite orientation shown by the green arrow. Due to this change in SG Index 2 sequencing directionality, sequences of 3rd Party Index 1 NEBNext indices should be reverse complemented on the SG-compatible Sample Sheet and placed in the Index 2 column of the SG compatible sample sheet. The 3rd party Index 2 should be listed in the Index 1 column of the SG compatible sample sheet. Since this index is read in the same orientation, no changes to the original index sequence are required.
ADJUSTING FOR INDEX LENGTH
The G4 sequencing platform has a minimum index length set at 8bps, so although the NEBNext Multiplex Oligos have an index length of 6bps, 8bps will need to be sequenced on the G4 Sequencing Platform. This minimum index length of 8bps will need to be accounted for on the SG compatible sample sheet.
For NEBNext Mulitplex Oligos Set 1, the modification to 8bps involves adding the two base pairs adjacent to the index, GT (See below for an example). For NEBNext Multiplex Oligos Set 2–4, the modification involves adding two bases (GT) adjacent to the index (see below for an example).
For Set 2–4, some of the NEBNext Indexes contain 8bps although only 6bps are reported in the NEBNext documentation. In this scenario, the index would be entered in the sample sheet as the reverse complement of the NEBNext 8bp Index Sequence. As an example, please see the table below for the SG Compatible sample sheet entries compared to the NEBNext reported sequences for Set 2. Similar to NEBNext Multiplex Oligos Set 1, there are other NEBNext Indexes within sets 2–4 which contain only the reported 6bps. With these indexes, the modification to 8bps requires adding the two bases (GT) adjacent to the index, similar to the modification for Set 1 Oligos.
MODIFYING THE SAMPLE SHEET
For NEBNext Multiplex Oligos (Index Primers Set 1):
- The SG Compatible Full Index Sequence for Index 1–12 will be the reverse complement of the reported 'expected index sequence read' as seen in the linked Product Insert + 'GT'
- For example, NEB Index 1:
- Expected: ATCACG
- SG Compatible: CGTGATGT
For NEBNext Multiplex Oligos (Index Primers Set 2):
- The SG Compatible Index Sequence for Index 13–23 will be the reverse complement of the NEBNext reported 'expected index sequence read' as seen in the linked NEBNext Product Insert + the two base pairs following the index
- For example, when entering as Index 2 in the SG Compatible Sample Sheet:
Please reach out to your FAS or Customer Care if you have any questions.
Version: 3 Apr 2024
