Use the information below in conjunction with those instructions and, in case of conflicts, follow the library prep manufacturer's instructions.
Quality Control Metric
Method
Interpretation
Quality of input DNA or RNA
Assess integrity (DIN or RIN score) and purity (OD 260/280 ratio)
Helps evaluate and control isolation method.
Size range before beginning library prep
BioAnalyzer, TapeStation, or equivalent method to visualize molecular weight
Helps determine an appropriate fragmentation method.
Size range after fragmentation
BioAnalyzer, TapeStation, or equivalent method to visualize size distribution
Assesses the efficiency of fragmentation and/or determine optimal SPRI bead: sample ratio.
Amount of library that is viable for sequencing
qPCR, QubIt, or equivalent method to assess the number of functional library molecules. Qubit is recommended to quantify libraries with different fragment size or broad fragment distribution.
Assesses ligation efficiency and determine the functionality of library.
Quality of the library
BioAnalyzer or TapeStation
Verify the absence of undesirable short fragments and determine optimal loading concentration.
Below are examples of what a trace for a clean library preparation should look like.
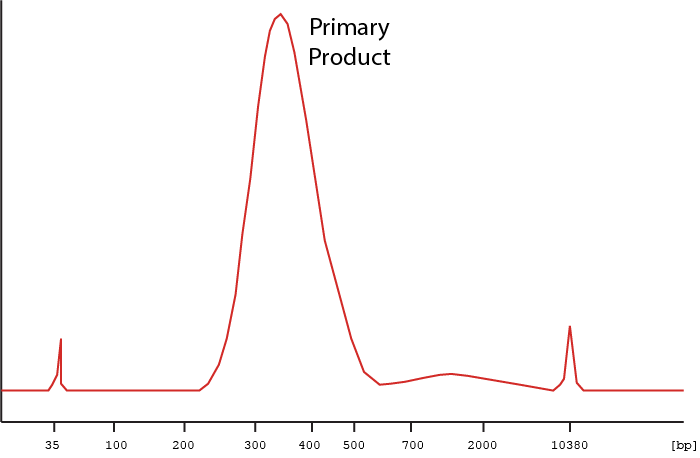
Figure 1 Example of a trace for a clean library preparation. Peaks at 35 and 10380 bp are size markers.
![]()
Figure 2 Example of a trace for a clean amplicon library preparation. Peaks at 35 and 10380 bp are size markers.
